Welcome to the Saier Lab
Our laboratory takes a multidisciplinary approach to science, using biochemical, molecular genetic, physiological, and bioinformatic approaches. We have three primary research interests, one concerned with transcriptional and metabolic regulation in bacteria, a second with transport protein evolution, and a third with the recently identified process of transposon-mediated directed mutation. We also created and now maintain the Transporter Classification DataBase (TCDB), which was adopted by the International Union of Biochemistry and Molecular Biology (IUBMB) as the primary source of information relating to molecular transport.
About us
Dr. Milton Saier is the head of the laboratory. The lab has two main branches: the wet lab (led by Dr. Zhongge Zhang) and the dry lab (led by Dr. Arturo Medrano-Soto). The progress of our experimental and bioinformatic projects depends on the work and interaction within our wonderfully eclectic Team of researchers, talented students, visiting scholars and external collaborators. For more details, please visit our Research and Publications pages. The software developed and maintained by our bioinformatics team is available in our GitHub repository and our web applications are available in our BioTools page, Please, do not hesitate to contact us If you have any questions/requests or if you would like to give us feedback.

Latest News
MSc degree obtained
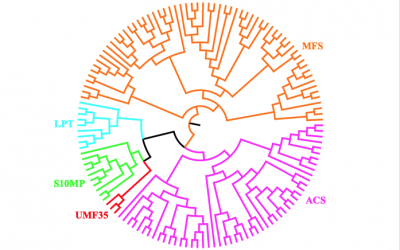
Congratulations to our grad student Sophia Beliaev for a successful defense of her Master's thesis on March 15. The title of her thesis is "Expansion of the Major Facilitator Superfamily (MFS) with the Sar TMS10 Membrane Protein (S10MP) Family."
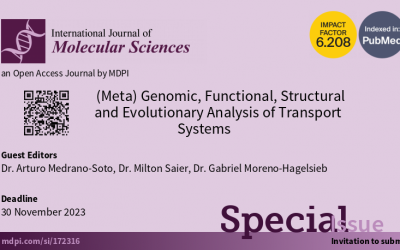
IJMS special issue
Drs. A Medrano-Sot, M Saier and G. Moreno-Hagelsieb are guest editors for an upcoming Special Issue to be published in the International Journal of Molecular Sciences (IJMS). The special issue's name is: (Meta)genomic, Functional, Structural and Evolutionary Analysis...
Hot off the press
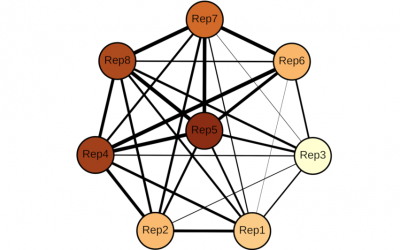
MSc. Kevin J Hendargo, collaborators Ashay O Patel, Onyeka S Chukwudozie, as well as Drs. Gabriel Moreno-Hagelsieb, J Andrés Christen, Arturo Medrano-Soto and Milton Saier just published a paper in the journal Microbial Physiology. The title is: Sequence Similarity...